
作者:Rubens Zimbres
编译:活水智能
最近,我发现了一个名为 neo4j-runway 的项目。Neo4j Runway 是一个 Python 库,简化了将关系数据迁移到图数据库的过程。它提供了与 OpenAI 交互的工具,用于数据发现和生成数据模型,还提供了生成代码和将数据加载到 Neo4j 实例中的工具。
换句话说,通过上传 CSV 文件,大模型将识别节点之间的关系,并自动生成知识图谱。
在健康医疗领域,知识图谱是一种强大的工具,用于组织和分析复杂的医疗数据。这些图谱能够以更易于理解的方式结构化信息,使得不同实体之间的关系更加清晰,例如疾病、治疗、患者和医疗提供者。
知识图谱在健康医疗行业中的应用
-
整合多样化的数据源:知识图谱可以整合来自各种来源的数据,如电子健康记录(EHRs)、医学研究论文、临床试验结果、基因组数据和患者历史记录。
-
改进临床决策:通过链接症状、诊断、治疗和结果,知识图谱可以增强临床决策支持系统(CDSS),因为它们考虑了大量相互关联的医学知识,可能提高诊断准确性和治疗效果。
-
个性化医疗:通过将患者特定数据与更广泛的医学知识相关联,促进制定个性化治疗计划。这包括理解基因信息、疾病机制和治疗反应之间的关系,从而提供更量身定制的医疗。
-
加速新药研发:在制药研究中,知识图谱可以通过识别潜在药物靶点和理解疾病涉及的生物路径,加速新药研发。
-
公共卫生与流行病学:有助于追踪疾病爆发、理解流行病学趋势和规划干预措施。因为它们可以整合来自各种公共卫生数据库、社交媒体和其他来源的数据,提供关于公共卫生威胁的实时洞察。
Neo4j Runway 简介
Neo4j Runway 是由 Alex Gilmore 创建的开源库。你可以在 GitHub 上找到代码库。
目前,该库仅支持使用 OpenAI 的大模型解析 CSV,并提供以下功能:
- 提取数据:利用大模型从数据中提取有意义的见解。
- 图数据建模:使用 OpenAI 和 Instructor Python 库开发准确的图数据模型。
- 数据摄取:利用 Runway 内置的 PyIngest,将数据加载到 Neo4j 中。
- 无需编写 Cypher 语句:大模型会完成所有工作。
本文除了演示用大模型把 CSV 文件转为知识图谱外,还使用了 Langchain 的 GraphCypherQAChain,用提示词生成 Cypher,无需编写一行 Cypher(用于查询 Neo4j 图数据库的 SQL 类语言)即可查询图谱。
该库提供了一个金融行业的示例,本文将测试在健康医疗场景中的效果。
数据准备
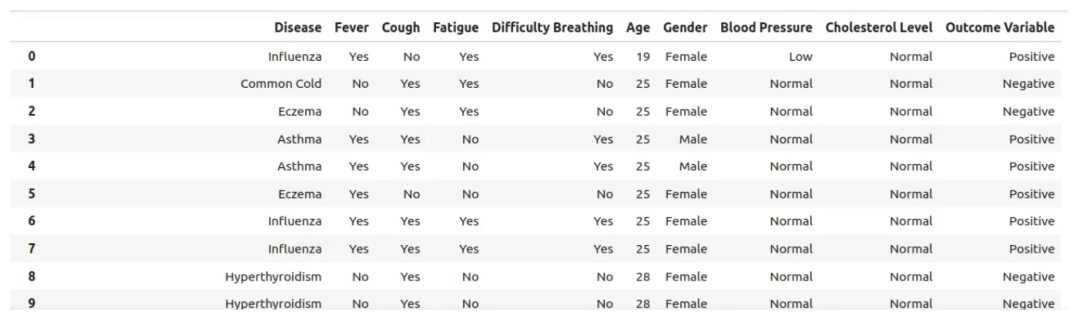
我们从 Kaggle 上下载了一个非常简单的数据集(疾病症状和患者档案数据集)。该数据集只有 10 列(疾病、发烧、咳嗽、疲劳、呼吸困难、年龄、性别、血压、胆固醇水平和结果变量),我们希望能够向大模型提供医疗报告,以获得诊断假设。
数据集下载链接:Kaggle 数据集
代码实现
1. 加载所需的库和环境变量
首先,加载所需的库:
sudo apt install python3-pydot graphviz
pip install neo4j-runway
然后,在 Python 中导入相关库:
import numpy as np
import pandas as pd
from neo4j_runway import Discovery, GraphDataModeler, IngestionGenerator, LLM, PyIngest
from IPython.display import display, Markdown, Image
加载环境变量:在 Neo4j Aura 中创建实例并进行身份验证。
load_dotenv()
OPENAI_API_KEY = os.getenv('sk-openaiapikeyhere')
NEO4J_URL = os.getenv('neo4j+s://your.databases.neo4j.io')
NEO4J_PASSWORD = os.getenv('yourneo4jpassword')
2. 加载医疗数据并整理格式
从 Kaggle 网站下载 CSV 文件,并将其加载到 Jupyter notebook 中。这个数据集非常简单,但对于测试概念非常有用。
disease_df = pd.read_csv('/home/user/Disease_symptom.csv')
disease_df
例如,我们可以创建一个列表,列出所有导致呼吸困难的疾病,这不仅对选择图中的节点很有趣,也有助于开发诊断假设:
disease_df[disease_df['Difficulty Breathing'] == 'Yes']
所有变量必须是字符串(库是这样设计的),即使是整数。然后,我们保存 CSV 文件:
disease_df.columns = disease_df.columns.str.strip()
for i in disease_df.columns:
disease_df[i] = disease_df[i].astype(str)
disease_df.to_csv('/home/user/disease_prepared.csv', index=False)
3. 用大模型识别重要数据元素
下一步是让大模型分析表格数据,识别对生成图数据模型重要的数据元素。
disc = Discovery(llm=llm, user_input=DATA_DESCRIPTION, data=disease_df)
disc.run()
这将生成数据分析的 Markdown 输出:

4. 创建初始模型
现在,让我们创建初始模型:
# 实例化图数据建模器
gdm = GraphDataModeler(llm=llm, discovery=disc)
# 生成模型
gdm.create_initial_model()
# 可视化数据模型
gdm.current_model.visualize()
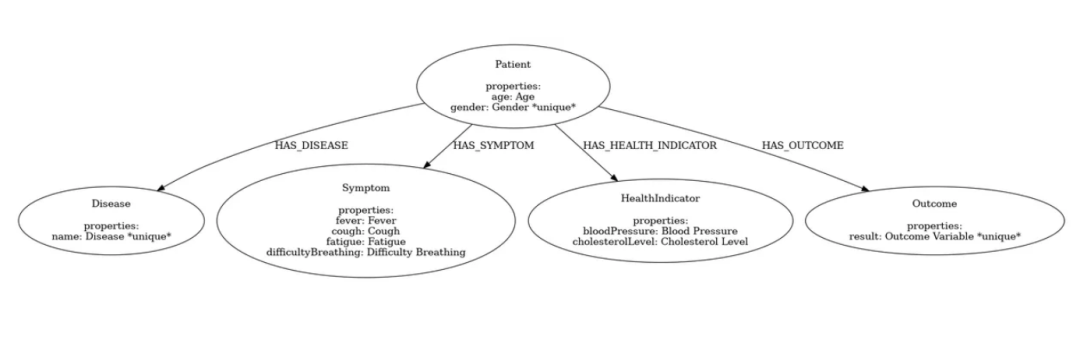
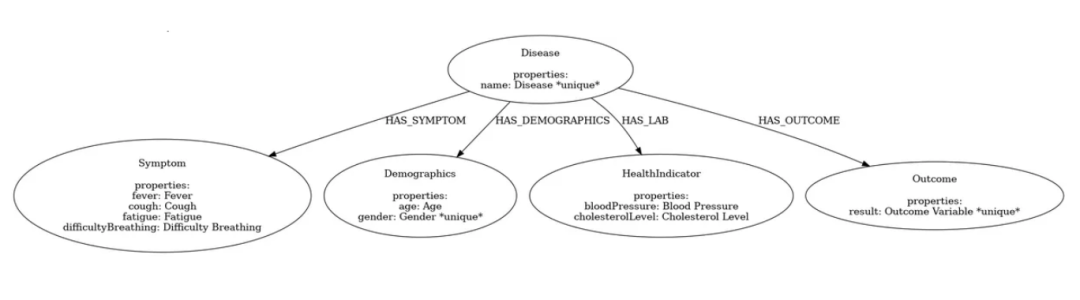
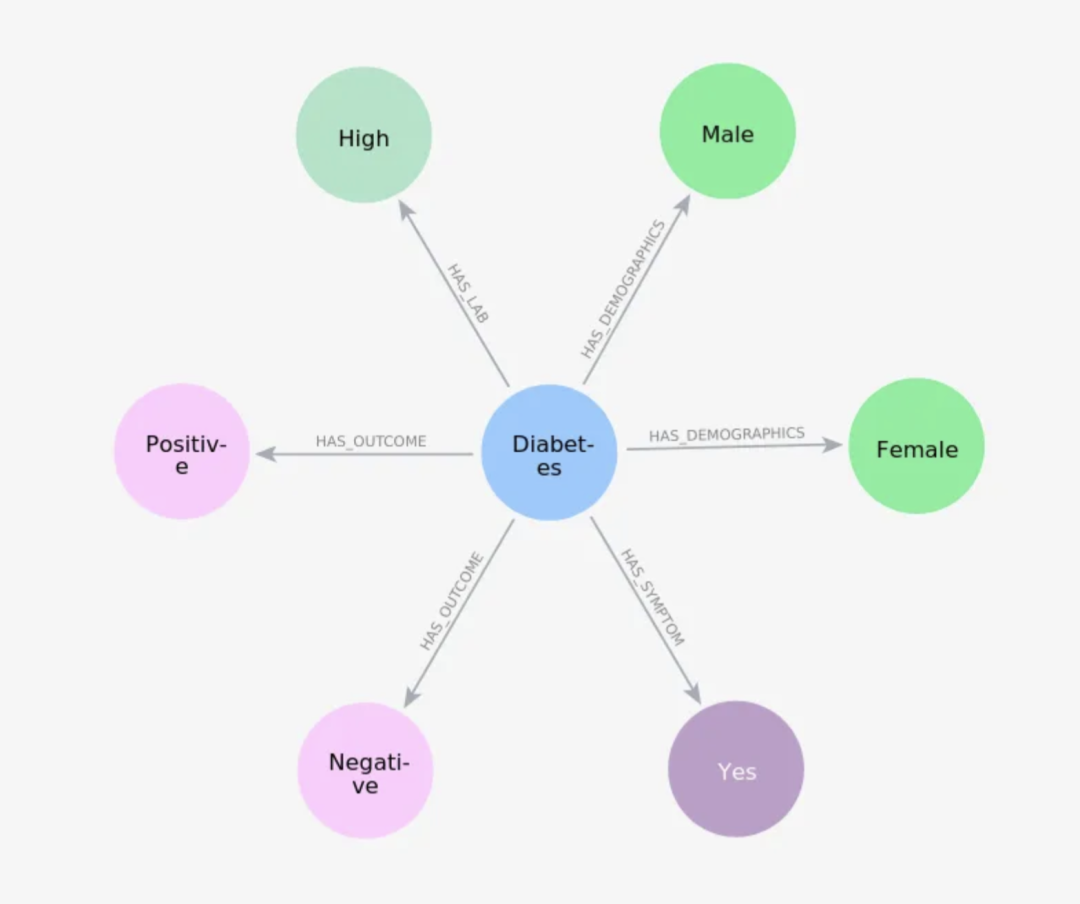
在这个模型中,我们的重点是疾病,所以我们将重新排列一些关系。
gdm.iterate_model(user_corrections='''
Let's think step by step. Please make the following updates to the data model:
1. Remove the relationships between Patient and Disease, between Patient and Symptom and between Patient and Outcome.
2. Change the Patient node into Demographics.
3. Create a relationship HAS_DEMOGRAPHICS from Disease to Demographics.
4. Create a relationship HAS_SYMPTOM from Disease to Symptom. If the Symptom value is No, remove this relationship.
5. Create a relationship HAS_LAB from Disease to HealthIndicator.
6. Create a relationship HAS_OUTCOME from Disease to Outcome.
''')
然后,加载并显示图像:
from IPython.display import Image, display
gdm.current_model.visualize().render('output', format='png')
img = Image('output.png', width=1200) # 调整宽度
display(img)
5. 用Neo4j生成图谱
现在我们可以生成 Cypher 代码和 YAML 文件,将数据加载到 Neo4j 中。
# 实例化数据摄取生成器
gen = IngestionGenerator(
data_model=gdm.current_model,
username="neo4j",
password='yourneo4jpasswordhere',
uri='neo4j+s://123654888.databases.neo4j.io',
database="neo4j",
csv_dir="/home/user/",
csv_name="disease_prepared.csv"
)
# 创建摄取 YAML
pyingest_yaml = gen.generate_pyingest_yaml_string()
# 保存本地 YAML 副本
gen.generate_pyingest_yaml_file(file_name="disease_prepared")
一切准备就绪后,让我们将数据加载到实例中:
PyIngest(yaml_string=pyingest_yaml, dataframe=disease_df)
进入 Neo4j Aura 实例,输入你的密码,并通过 Cypher 运行此查询:
MATCH (n)
WHERE n:Demographics OR n:Disease OR n:Symptom OR n:Outcome OR n:HealthIndicator
OPTIONAL MATCH (n)-[r]->(m)
RETURN n, r, m
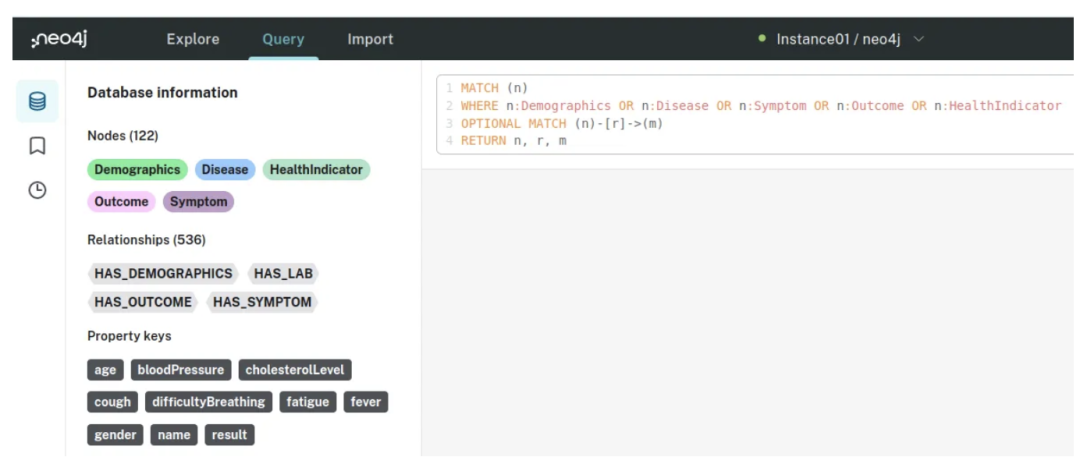
按下 CTRL + ENTER,你将看到以下结果:
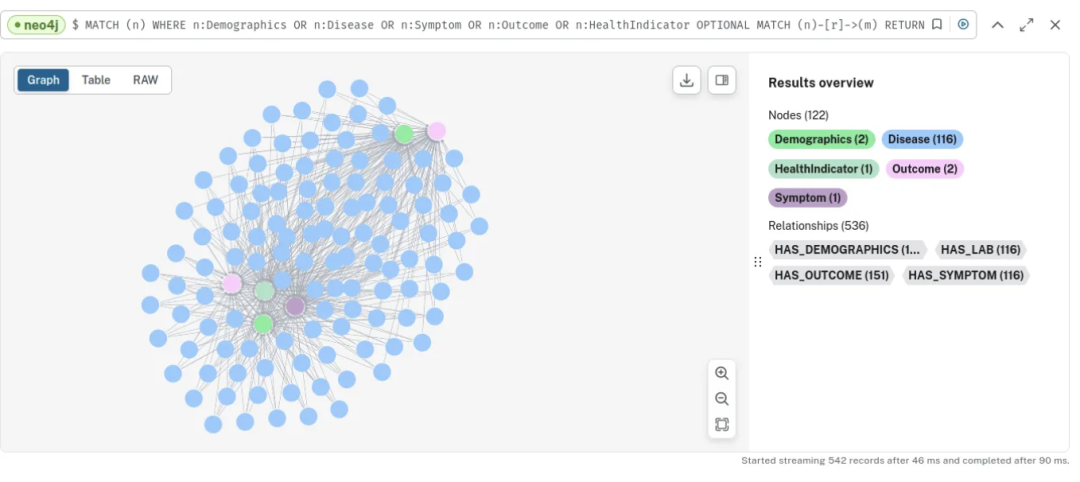
检查节点和关系后,我们发现症状、健康指标和人口统计数据之间有大量的相互连接:
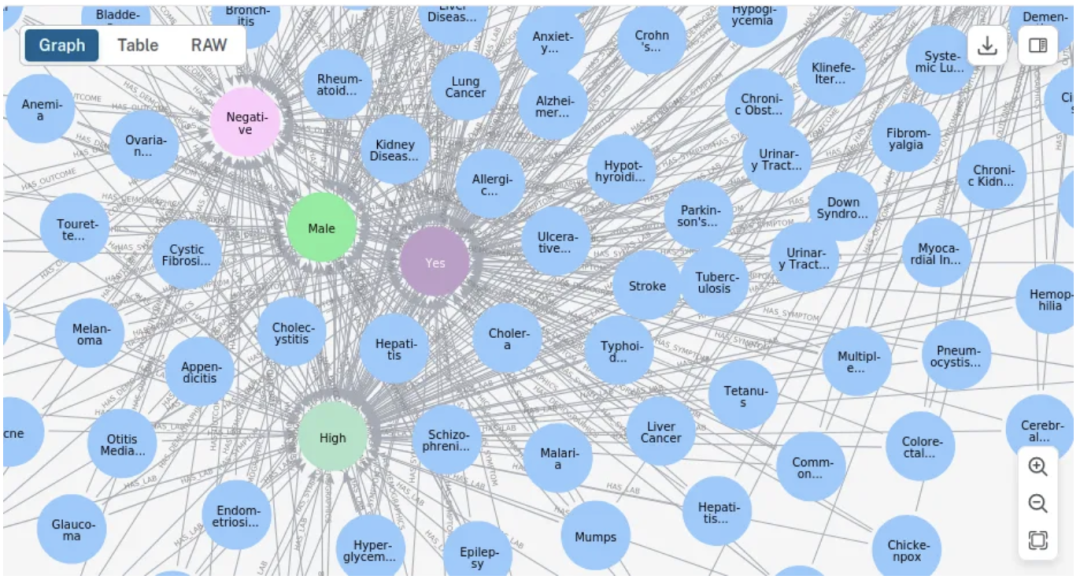
让我们看看糖尿病。由于没有应用过滤器,男性和女性都会出现,以及所有的实验室、人口统计和结果的可能性。
MATCH (n:Disease {name: 'Diabetes'})
WHERE n:Demographics OR n:Disease OR n:Symptom OR n:Outcome OR n:HealthIndicator
OPTIONAL MATCH (n)-[r]->(m)
RETURN n, r, m
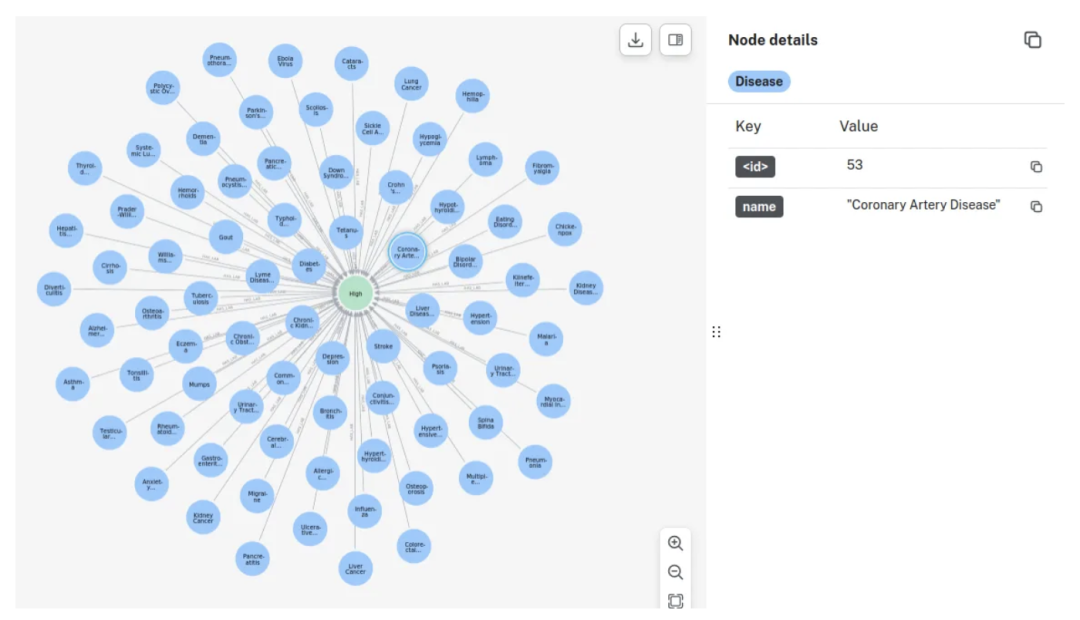
或者查看所有在临床检查中表现出高血压的疾病:
MATCH (d:Disease)
MATCH (d)-[r:HAS_LAB]->(l)
MATCH (d)-[r2:HAS_OUTCOME]->(o)
WHERE l.bloodPressure = 'High' AND o.result='Positive'
RETURN d, properties(d) AS disease_properties, r, properties(r) AS relationship_properties, l, properties(l) AS lab_properties
使用Langchain进行自动化查询
接下来我们向大模型(在本例中是 Google 的 Gemini-1.5-Flash)提交一份医疗报告,让它通过 Langchain(GraphCypherQAChain)自动生成 Cypher 查询,基于症状、健康指标等,返回患者可能患有的疾病。
1. 初始化环境
import warnings
import json
from langchain_community.graphs import Neo4jGraph
with warnings.catch_warnings():
warnings.simplefilter('ignore')
NEO4J_USERNAME = "neo4j"
NEO4J_DATABASE = 'neo4j'
NEO4J_URI = 'neo4j+s://1236547.databases.neo4j.io'
NEO4J_PASSWORD = 'yourneo4jdatabasepasswordhere'
从实例中获取知识图谱和模式:
kg = Neo4jGraph(
url=NEO4J_URI,
username=NEO4J_USERNAME,
password=NEO4J_PASSWORD,
database=NEO4J_DATABASE
)
kg.refresh_schema()
print(textwrap.fill(kg.schema, 60))
schema = kg.schema

2. 初始化 Vertex AI Gemini-1.5-Flash
from langchain.prompts.prompt import PromptTemplate
from langchain.chains import GraphCypherQAChain
from langchain.llms import VertexAI
# Initialize Vertex AI
vertexai.init(project="your-project", location="us-west4")
llm = VertexAI(model="gemini-1.5-flash")
3. 创建提示词
现在是最难的部分。为 Gemini-1.5-Flash 创建详细的指令,让它自动生成 Cypher 查询图数据库并获取我们需要的结果。
prompt_template = """
Let's think step by step:
Step 1: Task:
Generate an effective and concise Cypher statement with less than 256 characters to query a graph database.
Do not comment the code.
Step 2: Get to know the database schema: {schema}
Step 3: Instructions:
- In the cypher query, ONLY USE the provided relationship types and properties that appear in the schema AND in the user question.
- In the cypher query, do not use any other relationship types or properties in the user's question that are not contained in the provided schema.
- Regarding Age, NEVER work with the age itself. For example: 24 years old, use interval: more than 20 years old.
- USE ONLY ONE statement for Age, always use 'greater than', never 'less than' or 'equal'.
- DO NOT USE property keys that are not in the database.
Step 4: Examples:
Here are a few examples of generated Cypher statements for particular questions:
4.1 Which diseases present high blood pressure?
MATCH (d:Disease)
MATCH (d)-[r:HAS_LAB]->(l)
WHERE l.bloodPressure = 'High'
RETURN d.name
4.2 Which diseases present indicators as high blood pressure?
MATCH (d:Disease)
MATCH (d)-[r:HAS_LAB]->(l)
MATCH (d)-[r2:HAS_OUTCOME]->(o)
WHERE l.bloodPressure = 'High' AND o.result='Positive'
RETURN d, properties(d) AS disease_properties, r, properties(r) AS relationship_properties, l, properties(l) AS lab_properties
4.3 What is the name of a disease of the elderly where the patient presents high blood pressure, high cholesterol, fever, fatigue
MATCH (d:Disease)
MATCH (d)-[r1:HAS_LAB]->(lab)
MATCH (d)-[r2:HAS_SYMPTOM]->(symptom)
MATCH (symptom)-[r3:HAS_DEMOGRAPHICS]->(demo)
WHERE lab.bloodPressure = 'High' AND lab.cholesterolLevel = 'High' AND symptom.fever = 'Yes' AND symptom.fatigue = 'Yes' AND TOINTEGER(demo.age) >40
RETURN d.name
4.4 What disease gives you fever, fatigue, no cough, no short breathe in people with high cholesterol?
MATCH (d:Disease)-[r:HAS_SYMPTOM]->(s:Symptom)
WHERE s.fever = 'Yes' AND s.fatigue = 'Yes' AND s.difficultyBreathing = 'No' AND s.cough = 'No'
MATCH (d:Disease)-[r1:HAS_LAB]->(lab:HealthIndicator)
MATCH (d)-[r2:HAS_OUTCOME]->(o:Outcome)
WHERE lab.cholesterolLevel='High' AND o.result='Positive'
RETURN d, properties(d) AS disease_properties, r, properties(r) AS relationship_properties
Step 5. These are the values allowed for each entity:
- Fever: Indicates whether the patient has a fever (Yes/No).
- Cough: Indicates whether the patient has a cough (Yes/No).
- Fatigue: Indicates whether the patient experiences fatigue (Yes/No).
- Difficulty Breathing: Indicates whether the patient has difficulty breathing (Yes/No).
- Age: The age of the patient in years.
- Gender: The gender of the patient (Male/Female).
- Blood Pressure: The blood pressure level of the patient (Normal/High).
- Cholesterol Level: The cholesterol level of the patient (Normal/High).
- Outcome Variable: The outcome variable indicating the result of the diagnosis or assessment for the specific disease (Positive/Negative).
Step 6: Answer the question {question}.
"""
4. 设置 GraphCypherQAChain
cypher_prompt = PromptTemplate(
input_variables=["schema", "question"],
template=prompt_template
)
cypherChain = GraphCypherQAChain.from_llm(
VertexAI(temperature=0.1),
graph=kg,
verbose=True,
cypher_prompt=cypher_prompt,
top_k=10 # 这个值可以调整
)
5. 提交医疗报告
cypherChain.run("""
Patient Information:
Jane Doe, a 58-year-old female, was admitted on June 15, 2024.
Chief Complaint and History of Present Illness:
Jane reported a high fever up to 104°F, body pain, and a rash,
starting five days prior to admission.
Past Medical History:
Jane has no significant past medical history and no known allergies.
Physical Examination:
Jane's temperature was 102.8°F, heart rate 110 bpm, blood pressure 100/70 mmHg, and respiratory rate 20 breaths per minute. No petechiae or purpura were noted.
What disease may she have?
""")
输出结果:这里 Gemini-1.5-Flash 生成 Cypher 查询图数据库,通过 JSON 返回结果给 LLM,LLM 解释并返回可读的响应:
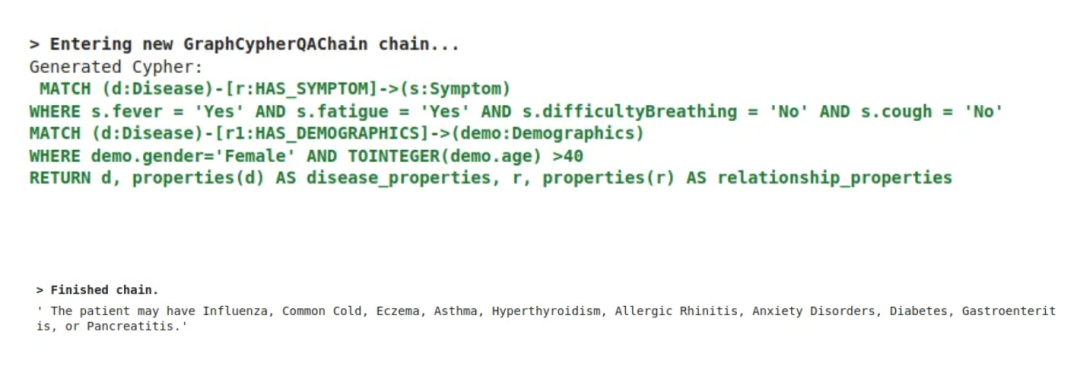
这个结果不考虑 Gemini-1.5-Flash 的知识库,只考虑它查询的知识图谱。想象一下,如果我们有一个包含 300 个特征的漂亮数据集!
注意,我们可以在 GraphCypherQAChain 中将 top_k 设置为 1 或其他值:

如果我们运行最后一个查询,我们将得到包含这些症状的 77 种疾病列表,但 top_k 设置为 1:
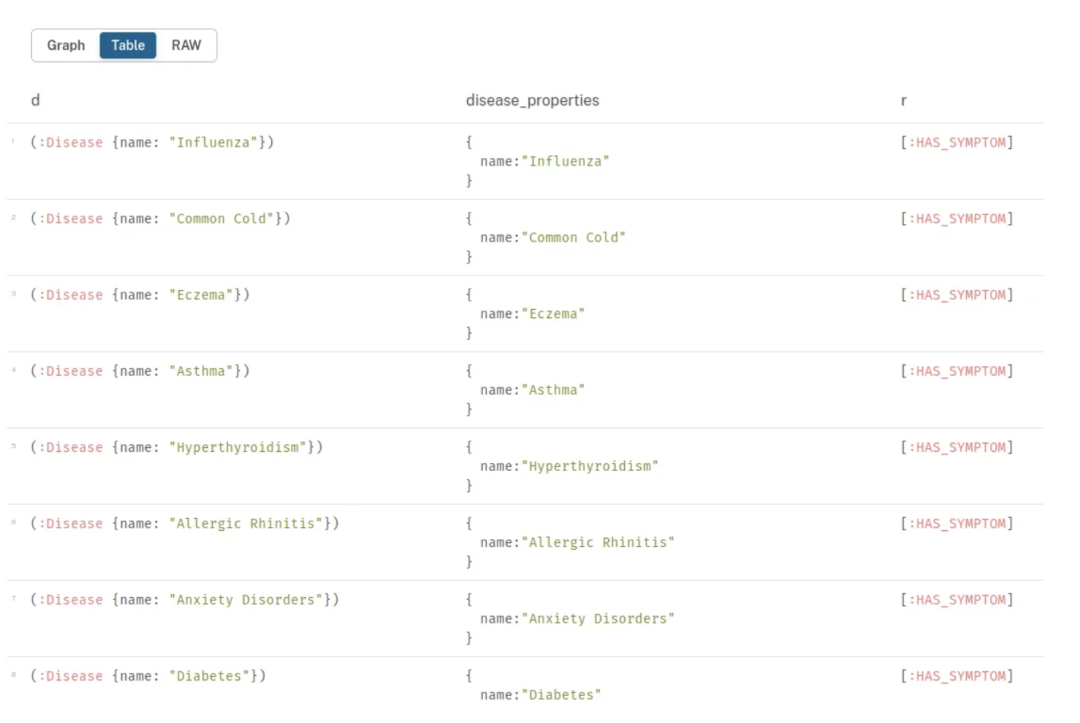
项目限制
需要注意的是,当前 neo4j-runway 项目还不成熟,具有以下限制:
- 仅支持单个 CSV 输入用于数据模型生成
- 节点只能有一个标签
- 仅支持唯一性和节点/关系键约束
- 关系不能有唯一性约束
- CSV 列引用相同节点属性在模型生成中不受支持
- 目前仅支持 OpenAI 模型
- Runway 包含的修改版 PyIngest 函数仅支持加载本地 Pandas DataFrame 或 CSV